Programming R like it's JavaScript / Python
R is the king of statistics languages for a good reason: There are hundreds of packages on CRAN for all sorts of analyses and the language is fluent for processing data in a variety of ways, especially when taking advantage of the hadleyverse packages. In my opinion, dplyr is the most beautiful library for manipulating data and ggplot2 is still the most flexible and elegant plotting library that exists in any programming language. Many libraries in other languages were inspired by or tried to imitate ggplot2 but no other library comes close to its elegance in day-to-day use.
While the R ecosystem is amazing, the R language unfortunately cannot compare to a true general-purpose language: Variable definitions and imports pollute the global namespace, and the only mechanisms available for structuring your code are to use source() for including another R file or importing R packages using library(). Both of these again pollute the global namespace. This makes R a messy solution for any mid-sized to large project.
Through my use of Python for everyday programming combined with excursions into web programming and using JavaScript, I got interested in how these other languages make their code more structured and reusable. After I while, I made a realization:
R becomes much more fun if you program it like JavaScript and Python!
This article will go over some of my techniques for using R against the way it was intended, starting from some minor stylistic changes to proposing different ways for structuring your code.
Assignment Operators
One of the idiosyncrasies of R that stick out most when coming to R from other programming languages is the <- assignment operator where other languages would use = for assignment. In R, both operators exist, but <- is preferred for historic reasons. Since except for subtle differences, the = and <- operators behave the same way, you should feel free to use whatever operator you are more comfortable with.
CommonJS-style Package Management
R and JavaScript share the problem that historically, both languages tended to define variables and functions into the default namespace (.GlobalEnv in R and window in JavaScript). While easy to use at first, importing additional modules will cause identifiers to conflict and variables to get overwritten. However, the JavaScript world found a solution to this problem in the CommonJS Modules/1.1 specification used for structuring node.js projects.
Put briefly, a node module is a .js file containing an exports variable. When a module is imported, all code in the .js file is executed and the value of the export variable returned. A simple example:
// file mymodule.js
var incrementAmount = 2;
exports.increment = function(a) {
return a + incrementAmount;
};
// file app.js
var myModule = require('./mymodule');
console.log(myModule.increment(3)); // prints 5
While this is a very simple mechanism, it allows for private variables in the modules (they simply don't get added to the exports object) and in the main script, only what is explicitly imported gets added to the namespace.
CommonJS Modules in R
Since R supports environments, it is possible to build the same strategy for structuring projects in R using my commonr library:
# file mymodule.R
increment.amount = 2
exports$increment = function(a) {
return a + increment.amount
}
# file app.R
library(commonr)
mymodule = require.r('./mymodule')
print(mymodule$increment(3)) # prints 5
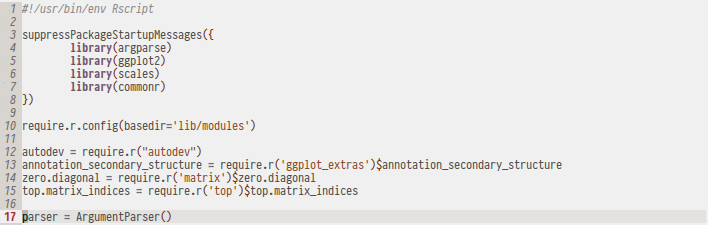
I've found that for my own projects, structuring code in commonr modules allowed me to cleanly compartmentalize and reuse code without having to spend time on creating a full R package. A nice example is my autodev module for automatically creating an R graphics device from a string.
Command Line Tools in R
A common workflow for R analyses is to split up different steps of the data processing pipeline into their own R scripts. While this is a clean way of separating an analysis, applying the same pipeline to different data will mean that the code has to be edited, leading to code duplication when applying the same pipeline to different data.
A cleaner solution would be to write our scripts in a way that they can be executed on different data sets with different parameters. Thanks to the Rscript binary, it is possible to program R scripts that behave like command line applications and the argparse module provides a powerful command line parser:
#!/usr/bin/env Rscript
suppressPackageStartupMessages({
library(ggplot2)
library(argparse)
})
parser = ArgumentParser()
parser$add_argument("in_data")
parser$add_argument("out_plotfile")
args = parser$parse_args()
df = read.table(args$in_data)
plt = ggplot(df, aes_auto()) + geom_point()
ggsave(args$out_plotfile, plt)
This is a plotting script that can be run from the command line to read a data file and produce a corresponding plot:
# assume that the script is in myscript.R
$ chmod +x myscript.R
$ ./myscript.R data.tsv data.svg
Conclusion
R is a powerful data analysis environment that can be made even better with a slightly creative workflow. I would be curious to see what you think about these suggestions and what other tweaks you have come up with!